Safety of Aquatic Animals as Human Protein Sources amid SARS-CoV-2 Concerns
Published 10 January, 2024
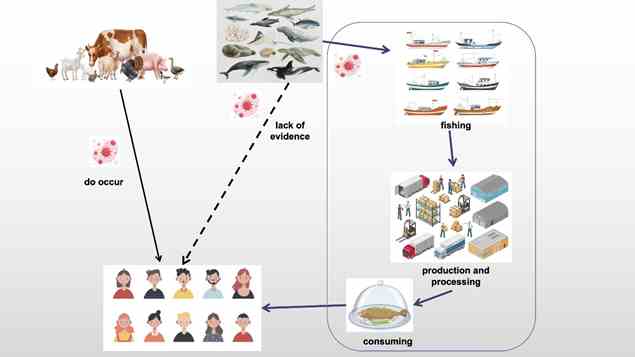
Aquatic animals have historically constituted a vital and nutritious dietary component for humans, contributing to nearly 20% of animal protein intake for approximately 3.3 billion people. Unlike terrestrial animals, there has been no evidence indicating that aquatic animals serve as reservoirs for zoonotic viruses. However, multiple cases of SARS-CoV-2 from cold-chain aquatic food and environmental samples have evoked worldwide concerns, despite the incidence being notably lower than that associated with poultry and livestock.
In a recent study published in the KeAi journal Reproduction and Breeding, a team of researchers from China and Canada developed a virus-mining pipeline to evaluate the risk of infections by aquatic animals.
The initial phase involved screening publicly available databases to gather transcriptomic and genomic data from commonly consumed aquatic species. This effort resulted in the acquisition of RNA-seq libraries of 70 aquatic species and reference genomes of 55 aquatic species. Human respiratory and intestine-related virus genomic sequences, such as coronavirus and influenza virus. were downloaded and used to build a virus reference genome pool. Two strategies were adopted to map the aquatic animals’ transcriptomes onto the genomes of human respiratory- and intestine-related viruses.
“Positive hints occurred in all positive control groups, which confirms the reliability of our pipeline,” explains senior and co-corresponding author Jing Luo, a professor in State Key Laboratory for Conservation and Utilization of Bio-resource at Yunnan University. “Besides, both mapping strategies identified fragments of Influenza A Virus from a salmon skin sample. This fragment could result from either contamination during sampling, or an IAV infection of fish, but no similar viral fragment exists in the transcriptomes of other tissues of the same S. salar sample.”
The researchers recovered and verified this human-associated viral fragment through de novo assembly and phylogenetic analysis. The results show high homology between the fragment and human IAV (H7N9).
“We believe that the fragment most likely came from sample contamination by human handling rather than viral infection,” adds Luo. “Apart from this false-positive result, analyses fail to find any human-associated viruses in the other aquatic animal transcriptomes.”
Multiple recent cases of SARS-CoV-2 have shown that cold-chain food and environmental contamination play a role in the spread of SARS-CoV-2, but through human contamination. In addressing that, the team concluded that aquatic animals are safe sources of protein for humans, albeit under the caveat of safe processing and storage. “Therefore, the processing of frozen aquatic animal products is critical to controlling the spread of the virus, and this should be carefully monitored,” said Luo.
Contact author details: Jing Luo, State Key Laboratory for Conservation and Utilization of Bio-resource, School of Ecology and Environment and School of Life Sciences, Yunnan University, Kunming, 650091, Yunnan, China. jingluo@ynu.edu.cn
Funder: This work was supported by the National Natural Science Foundation of China (grant no. U1902201, 32293253), Yunnan Fundamental Research Projects (grant no. 202301BF070001-019) and National Key R&D Program of China (grant no. 2022YFC260250302).
Conflict of interest: The authors declare the following financial interests/personal relationships which may be considered potential competing interests: Jing Luo is an editorial board member for Reproduction and Breeding and was not involved in the editorial review or the decision to publish this article. All authors declare that there are no competing interests.
See the article: Yuan Chen., et al., Transcriptome analysis confirms aquatic animals have less risk by carrying on human respiratory viruses, Reproduction and Breeding, Volume 3, Issue 4, 2023, Pages 161-168. https://doi.org/10.1016/j.repbre.2023.09.002

