Disentangling the influence of evolution and environment on plant traits
Published 03 September, 2025
Comparative analysis has long served as a foundational approach in ecological and evolutionary sciences, providing insights into the mechanisms underlying biodiversity patterns. However, trait similarities among species often result not only from shared ecological conditions but also from common ancestry.
To that end, phylogenetic generalized linear models (PGLMs) have been increasingly used in ecological research, particularly in investigating trait–environment relationships, improving species distribution models, and exploring the evolutionary dynamics of ecological strategies. PGLMs incorporate phylogenetic relationships by embedding a phylogenetic covariance matrix in the error structure, enabling the analysis of continuous or binary response variables while accounting for evolutionary relatedness among taxa.
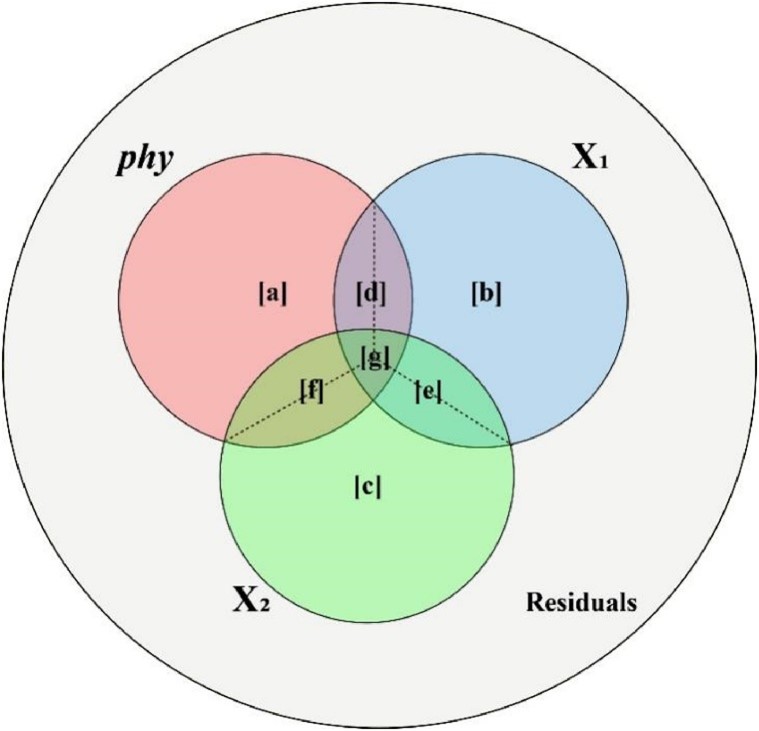
Despite its utility, the partial R2 framework has notable limitations. The sum of partial R2 values for all predictors (including phylogeny) often fails to equal the total R2 of the model. This discrepancy arises from the non-additive nature of explained variance when predictors are correlated, a well-known issue in regression analysis.
In an article published in the KeAi journal Plant Diversity, Dr. Lai and his team at Nanjing Forestry University in China present their new R package, phylolm.hp. This package extends the "averaged shared variance" (ASV) concept to the PGLM framework.
“The ASV method overcomes multicollinearity effects by fairly distributing overlapping explained variance among correlated predictors, achieving more transparent quantification of each variable's contribution,” explains Lai. “Specifically, the phylolm.hp package can calculate likelihood-based individual R² contributions for phylogeny and each predictor while considering both unique and shared explained variance. This feature makes it more accurate than traditional partial R² methods when handling correlated predictors.”
The development builds on a successful series of related tools. The research team has previously created several widely adopted R packages, including rdacca.hp (cited over 800 times), glmm.hp (more than 300 citations), and gam.hp (approximately 30 citations as of June 2025) . This growing usage highlights the broad recognition of the method's utility in advancing statistical modeling and data interpretation in ecological studies.
"This methodological innovation enriches the statistical toolkit for comparative biology and phylogenetic analysis, offering researchers more nuanced insights into the ecological and evolutionary drivers of trait variation,” adds Lai.
Funder: This research was supported by the National Natural Science Foundation of China (32271551, 32371603), National Key Research and Development Program of China (2023YFF0805800) and the Metasequoia funding of Nanjing Forestry University.
Conflict of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Article title: Evaluating the relative importance of phylogeny and predictors in phylogenetic generalized linear models using the phylolm.hp R package, Plant Diversity, https://doi.org/10.1016/j.pld.2025.06.003