Researchers shine new light on how urea denatures protein
Published 09 December, 2022
Urea is an organic compound that is found in abundant levels in urine. It is a potent denaturant – in other words, it has the ability to modify the molecular structure of proteins. Urea unfolds the proteins by disrupting their three-dimensional (3D) structure and native function. However, how it does this is not fully understood. Extensive simulations lend support to both direct and indirect interaction models, so a team of researchers from Singapore embarked on a study in an attempt to settle the question.
Their results, published in the KeAi journal Magnetic Resonance Letters, provide strong evidence that urea causes protein unfolding and denaturation via a series of direct, but transient interactions.
The team used two-dimensional (2D) nuclear magnetic resonance (NMR) to investigate the effect of low urea concentrations on the human intestinal fatty acid binding protein (hIFABP).
“The 3D conformation of hIFABP is well-resolved and its stability, even in the presence of denaturants such as urea, makes it an ideal candidate for understanding how proteins begin to unfold,” explains Wong Zhi Wei, one of the authors from National University of Singapore. Using hydrogen-deuterium exchange (HDX), where hydrogens are replaced with stable deuterium isotopes, urea was found to destabilise hIFABP non-uniformly and it “attacked” stable residues protected from HDX.
By assessing trends in signal intensity fluctuations with changing urea concentration, the scientists found that different amino acids in hIFABP were affected by urea disproportionately and were dependent on their local structures. According to Wei, “Together, these demonstrate that urea interacts with specific residues – providing concrete evidence to support a direct model.”
To understand how urea exerts its denaturing effects, researchers first identified water molecules important for stability that were tightly bound to hIFABP. At low concentrations, they found urea could not disrupt these robust interactions. Furthermore, no firm binding of urea directly to hIFABP was observed.
“From our results, we conclude that while urea interacts in a direct manner, these associations are not strong and are transient in nature”, explains Yang Daiwen, Professor of Structural Biology at National University of Singapore. He adds: “However, our methods do not preclude the possibility that urea has subtle and weak contacts with protein surfaces”.
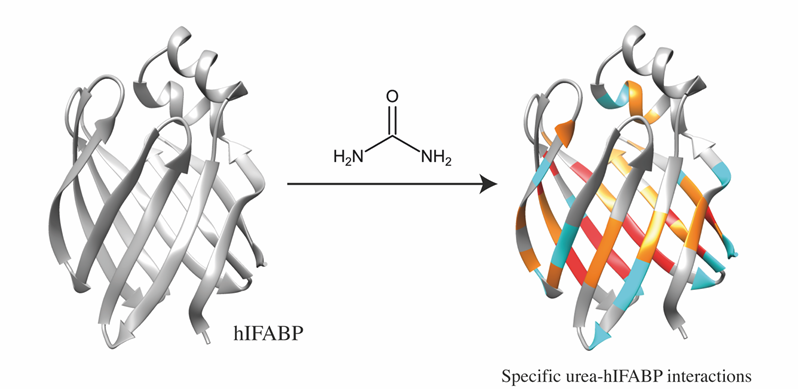
IMAGE: Protein regions involved in interactions with urea. Credit: the authors.
###
Contact the corresponding author: Daiwen Yang, dbsydw@nus.edu.sg

